
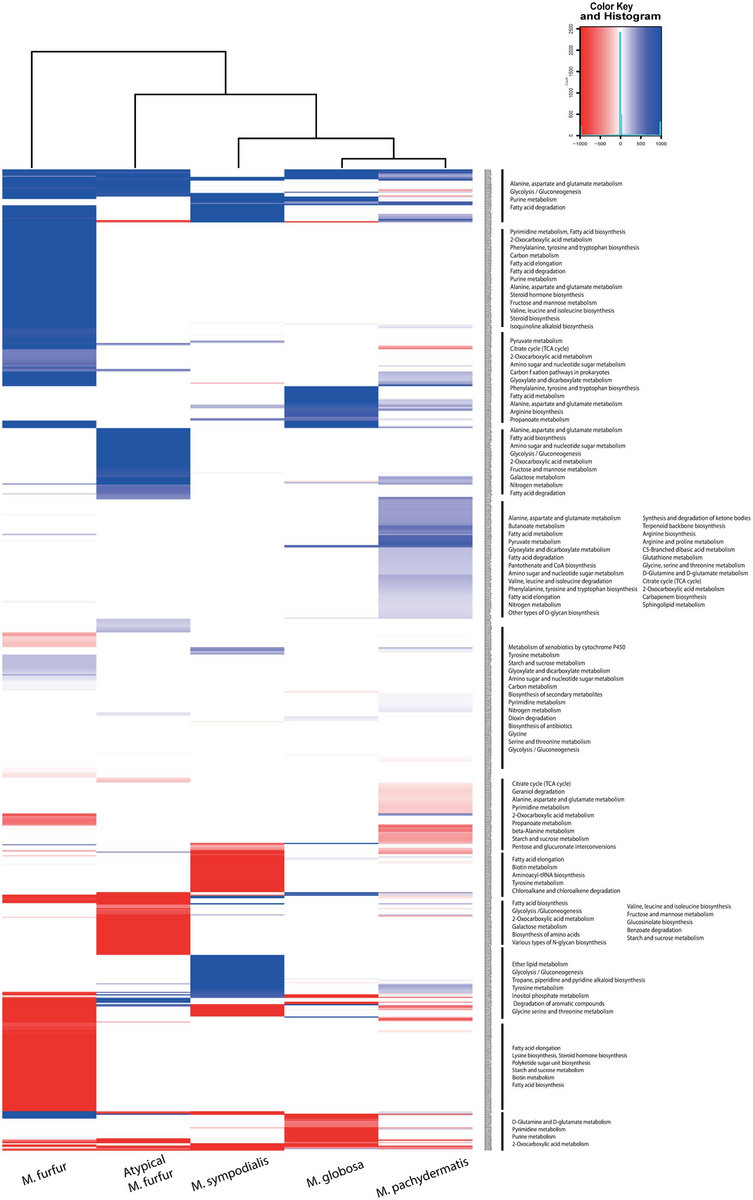
HTS enables the untargeted acquisition of extremely large amounts of sequence data from diverse sample types and thus represents an ideal and unique solution for the generic detection of highly diverse viruses. High-throughput sequencing (HTS) technologies have become an integral part of research and diagnostics toolbox in life sciences, including phytopathology and plant virology. By presenting the bioinformatic tools and a detailed overview of the consecutive steps that can be used to implement a well-structured HTS data analysis in an easy and accessible way, this paper is targeted at both beginners and expert scientists engaging in HTS plant virome projects. We start from sample preparation and nucleic acid extraction as appropriate to the chosen HTS strategy, which is followed by basic data analysis requirements, an extensive overview of the in-depth data processing options, and taxonomic classification of viral sequences detected. Here, we present a critical overview of the steps involved in HTS as employed for plant virus detection and virome characterization.
As HTS technologies are heavily relying on bioinformatics analysis of the huge amount of generated sequences, it is of utmost importance that researchers can rely on efficient and reliable bioinformatic tools and can understand the principles, advantages, and disadvantages of the tools used. We demonstrate that STEAK is a robust tool, which allows analysts to flexibly detect and evaluate TE and retroviral integrations in a diverse range of sequencing projects for both research and clinical purposes.High-throughput sequencing (HTS) technologies have become indispensable tools assisting plant virus diagnostics and research thanks to their ability to detect any plant virus in a sample without prior knowledge.
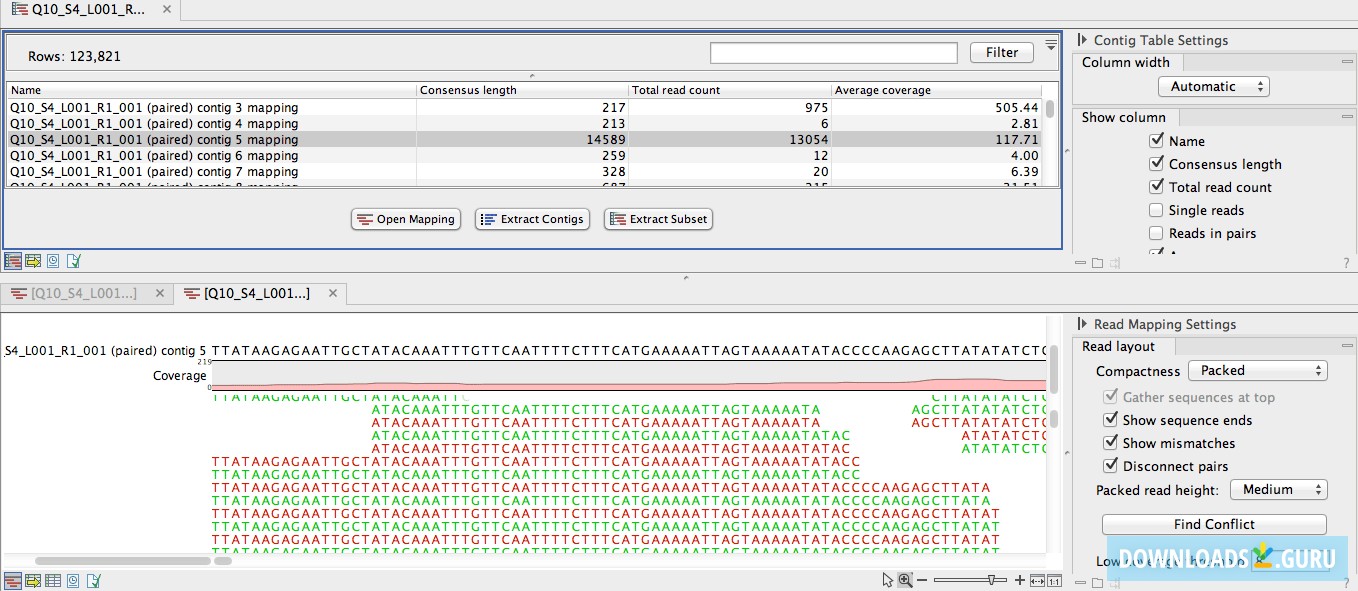
Allocating more memory into clc genomics workbench software#
We show that STEAK outperforms other software in terms of computational efficiency, sensitivity, and specificity. We highlight the capabilities of STEAK by comparing its efficacy in locating HERV-K HML-2 in clinical whole genome projects, target enrichment sequences, and in the 1000 Genomes CEU Trio to the performance of other TE and virus detecting tools. Here we describe our program STEAK, a massively parallel software designed to detect chimeric reads in high-throughput sequencing data for a broad number of applications such as identifying presence/absence, as well as discovery of transposable elements (TEs), and retroviral integrations. Even though numerous software have been developed to make sense of large genomics datasets, a major short falling of these has been the inability to cope with repetitive regions, specifically to validate structural variants and accordingly assess their role in disease. The advancements of high-throughput genomics have unveiled much about the human genome highlighting the importance of variations between individuals and their contribution to disease. We rescue a rare ten-nucleotide frameshift deletion in CR1, a top Alzheimer's disease gene, found in disease cases but not in controls. We present an algorithm to resolve most camouflaged regions and apply it to the Alzheimer's Disease Sequencing Project.
Linked-read or long-read sequencing technologies from 10x Genomics, PacBio, and Oxford Nanopore Technologies reduce dark protein-coding regions to approximately 50.5%, 35.6%, and 9.6%, respectively. We identify dark regions that are present in protein-coding exons across 748 genes. Of these gene bodies, 8.7% are completely dark and 35.2% are ≥ 5% dark. Results: Based on standard whole-genome Illumina sequencing data, we identify 36,794 dark regions in 6054 gene bodies from pathways important to human health, development, and reproduction. We assess how well long-read or linked-read technologies resolve these regions. Here, we identify regions with few mappable reads that we call dark by depth, and others that have ambiguous alignment, called camouflaged. Background: The human genome contains "dark" gene regions that cannot be adequately assembled or aligned using standard short-read sequencing technologies, preventing researchers from identifying mutations within these gene regions that may be relevant to human disease.


 0 kommentar(er)
0 kommentar(er)
